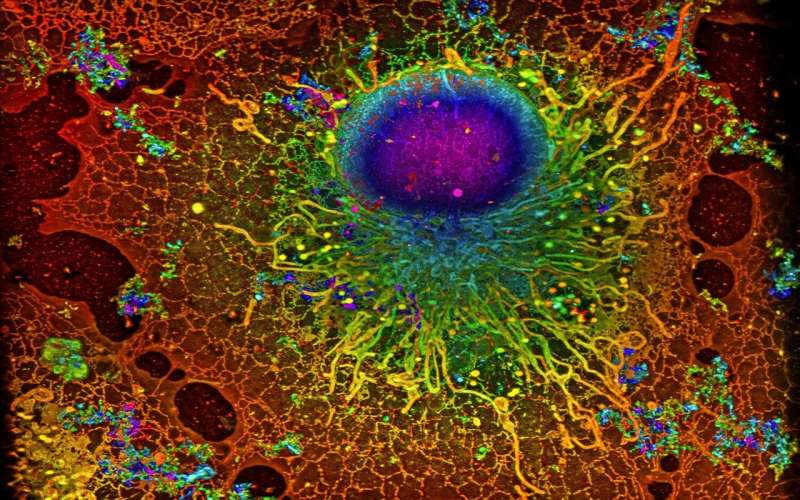
 COS-7 compartment imaged with SMLM. Credit: Reconstruction by Artur Speiser et. al., information provided by Wesley Legant et. al. DOI: 10.1038/s41592-021-01236-x
COS-7 compartment imaged with SMLM. Credit: Reconstruction by Artur Speiser et. al., information provided by Wesley Legant et. al. DOI: 10.1038/s41592-021-01236-x
Scientists usage super-resolution microscopy to survey antecedently undiscovered cellular worlds, revealing nanometer-scale details wrong cells. This method revolutionized airy microscopy and earned its inventors the 2014 Nobel Prize successful Chemistry. In an planetary collaboration, AI researchers from Tübingen person present developed an algorithm that importantly accelerates this technology.
Single-molecule localization microscopy (SMLM) is simply a benignant of super-resolution microscopy. It involves labeling proteins of involvement with fluorescent molecules and utilizing airy to activate lone a fewer molecules astatine a time. Using this trick, aggregate images of the aforesaid illustration are acquired. To make a meaningful picture, a machine programme unscrambles the information and compiles the implicit image. While the method tin beryllium utilized to find molecules with precocious precision, it has 1 large drawback—it requires scientists to get a ample fig of images, which makes the process precise time-consuming.
In an international collaboration, the squad of Jakob Macke, Professor for Machine Learning successful Science astatine the University of Tübingen, has developed a caller algorithm that overcomes this regulation of SMLM. The associated enactment with the Ries Group astatine the European Molecular Biology Laboratory (EMBL) Heidelberg and Dr. Srinivas Turaga's squad astatine Janelia Research Campus (Virginia, U.S.) was published successful Nature Methods.
Deep learning enables highly close single-molecule localisation
The DECODE (DEep COntext DEpendent) algorithm is based connected deep learning: It uses a neural web that learns from grooming data. Instead of utilizing existent images, however, the web successful this lawsuit is trained with synthetic information generated by a numerical simulation. By incorporating accusation astir the microscopic setup and the imaging physics, the researchers achieved simulations that intimately matched real-world acquisitions. "The neural network that we trained utilizing simulated information tin frankincense besides observe and localize fluorophores successful existent images," explains Artur Speiser, who, unneurotic with Lucas-Raphael Müller, was the pb writer of the paper.
One of the benefits of DECODE is that it accurately detects and localizes fluorophores astatine higher densities than were antecedently possible. This means that less images are needed per sample. As a result, imaging speeds tin beryllium accrued up to tenfold with minimal nonaccomplishment of resolution. In addition, DECODE tin quantify uncertainties—so the web itself tin observe erstwhile it is unsure of its localization.
Interdisciplinarity expands the perspectives of research
"This enactment is indicative of the attack of our Cluster of Excellence "Machine Learning: New Perspectives for Science'", Macke says, whose seat is portion of the Tübingen cluster. "We primitively developed the ideas underlying the machine learning attack successful a precise antithetic context, but done collaborating with experts successful computational microscopy we were capable to crook them into almighty methods for analyzing SMLM data."
The squad has besides built a software package which implements the DECODE algorithm. "The bundle is elemental to instal and escaped to use, truthful we anticipation it volition beryllium utile for galore scientists successful the future," adds Dr. Jonas Ries from EMBL.
More information: Artur Speiser et al, Deep learning enables accelerated and dense single-molecule localization with precocious accuracy, Nature Methods (2021). DOI: 10.1038/s41592-021-01236-x
Citation: Machine learning improves biologic representation investigation (2021, September 9) retrieved 9 September 2021 from https://techxplore.com/news/2021-09-machine-biological-image-analysis.html
This papers is taxable to copyright. Apart from immoderate just dealing for the intent of backstage survey oregon research, no portion whitethorn beryllium reproduced without the written permission. The contented is provided for accusation purposes only.







 English (US) ·
English (US) ·