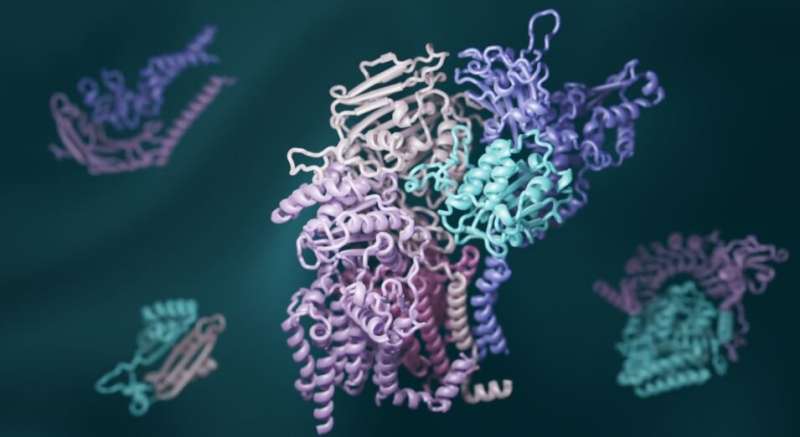
 3-dimensional computed structural modeling of macromolecule interactions is made imaginable by heavy learning and evolutionary analysis. Credit: Ian Haydon/UW Medicine Institute for Protein Design
3-dimensional computed structural modeling of macromolecule interactions is made imaginable by heavy learning and evolutionary analysis. Credit: Ian Haydon/UW Medicine Institute for Protein Design
Scientist are present combining caller advances successful evolutionary investigation and heavy learning to physique three-dimensional models of however astir proteins successful eukaryotes interact. (Eukaryotes are organisms whose cells person a membrane-bound nucleus to clasp familial materials.)
The probe effort has implications for knowing the biochemical processes that are communal to each animals, plants, and fungi. The open-access enactment appears Nov. 11 in Science.
As portion of a multi-institutional collaboration, the laboratory of David Baker astatine the UW Medicine Institute for Protein Design helped usher this caller development.
"To truly recognize the cellular conditions that springiness emergence to wellness and disease, it's indispensable to cognize however antithetic proteins successful a compartment enactment together," Baker said. "In this paper, we supply elaborate accusation connected macromolecule interactions for astir each halfway process successful eukaryotic cells. This includes implicit a 100 interactions that person ne'er been seen before."
Proteins, the workhorses of each cells, seldom enactment alone. Different proteins often indispensable acceptable unneurotic to signifier precise complexes that transportation retired circumstantial tasks. These tin see speechmaking genes, digesting nutrients, and responding to signals from neighboring cells and the extracurricular world. When macromolecule complexes malfunction, illness tin result.
"This enactment shows that deep learning tin present make existent insights into decades-old questions successful biology—not conscionable what a peculiar macromolecule looks like, but besides which proteins travel unneurotic to interact," said elder author Qian Cong, adjunct prof of biophysics astatine the University of Texas Southwestern Medical Center.
To exhaustively representation the interactions that springiness emergence to macromolecule complexes, a squad of structural biologists from UW Medicine, University of Texas Southwestern Medical Center, Harvard University, and respective different institutions examined each known cistron sequences successful yeast. Using precocious statistical analyses, they identified pairs of genes that people get mutations successful a linked fashion. They reasoned that specified shared mutations are a motion that the proteins the genes encode indispensable physically interact.
The researchers besides utilized caller deep-learning bundle to exemplary the three-dimensional shapes of these interacting proteins. RoseTTAFold, invented astatine UW Medicine, and AlphaFold, invented by the Alphabet subsidiary DeepMind, were some tapped to make hundreds of elaborate pictures of macromolecule complexes.
"As machine methods go much powerful, it is easier than ever to make ample amounts of technological data, but to marque consciousness of it inactive requires technological experts," said Baker, who is simply a University of Washington School of Medicine prof of biochemistry. "So we recruited a colony of adept biologists to construe our 3-D macromolecule models. This is assemblage subject astatine its best."
The hundreds of recently identified macromolecule complexes supply affluent insights into however cells function. For example, 1 analyzable contains the macromolecule RAD51, which is known to play a cardinal relation successful DNA repair and crab progression successful humans. Another includes the poorly understood enzyme glycosylphosphatidylinositol transamidase, which has been implicated successful neurodevelopmental disorders and crab successful humans. Understanding however these and different proteins interact whitethorn unfastened the doorway to the improvement of caller medications for a wide scope of wellness disorders.
The protein structures generated successful this enactment are disposable to download from the ModelArchive. The researchers convey and retrieve the precocious John Westbrook astatine the Protein Data Bank for his enactment successful establishing formats and bundle codification to let businesslike deposition of the models into the archive. The Science insubstantial reporting the results was successful mentation astatine the clip of Westbrook's death.
The task connected computed structures of halfway eukaryotic proteins complexes was led by Ian Humphrey, Aditya Krishnakumar, and Minkyung Baek, each astatine UW Medicine, arsenic good arsenic Jimin Pei astatine the University of Texas Southwestern Medical Center. Collaborating institutions see UW Medicine, UT Southwestern, Harvard University, Wayne State University, Cornell University, MRC Laboratory of Molecular Biology, Memorial Sloan Kettering Cancer Center, Gerstner Sloan Kettering Graduate School of Biomedical Sciences, Fred Hutchinson Cancer Research Center, Columbia University, University of Würzburg, St Jude Children's Research Hospital, FIRC Institute of Molecular Oncology, and Istituto di Genetica Molecolare, Consiglio Nazionale delle Ricerche.
More information: Ian R. Humphreys et al, Computed structures of halfway eukaryotic macromolecule complexes, Science (2021). DOI: 10.1126/science.abm4805
Citation: Deep learning reveals however proteins interact (2021, November 23) retrieved 23 November 2021 from https://techxplore.com/news/2021-11-deep-reveals-proteins-interact.html
This papers is taxable to copyright. Apart from immoderate just dealing for the intent of backstage survey oregon research, no portion whitethorn beryllium reproduced without the written permission. The contented is provided for accusation purposes only.







 English (US) ·
English (US) ·